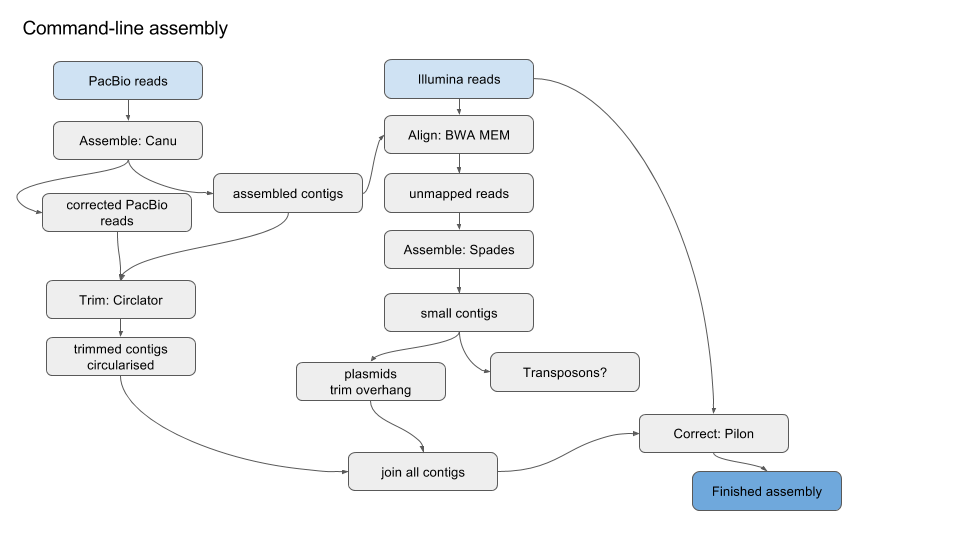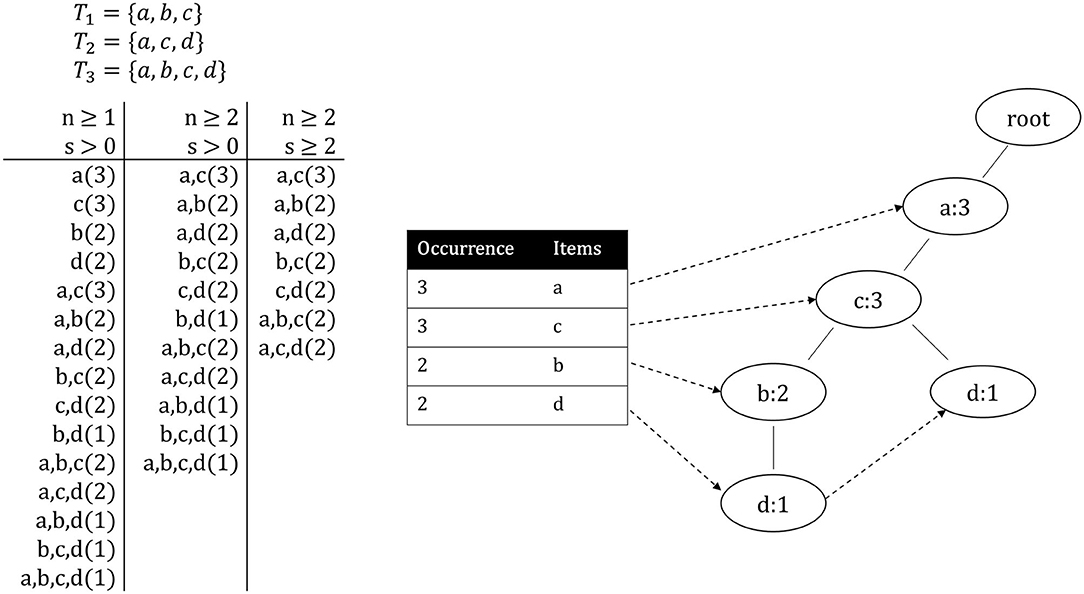
PDF) HGA: De novo genome assembly method for bacterial genomes using high coverage short sequencing reads
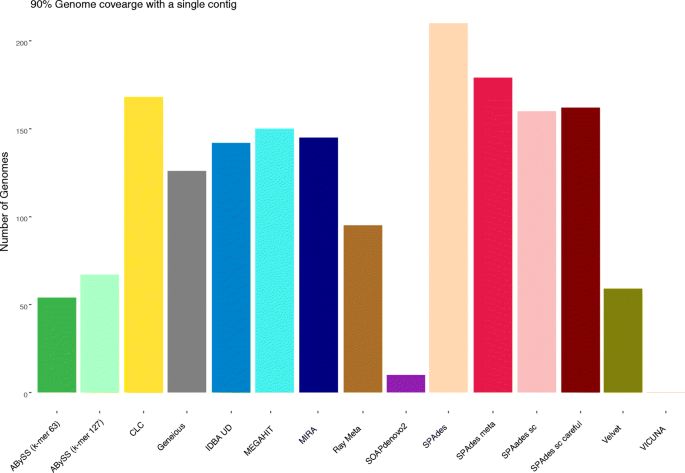
Choice of assembly software has a critical impact on virome characterisation | Microbiome | Full Text

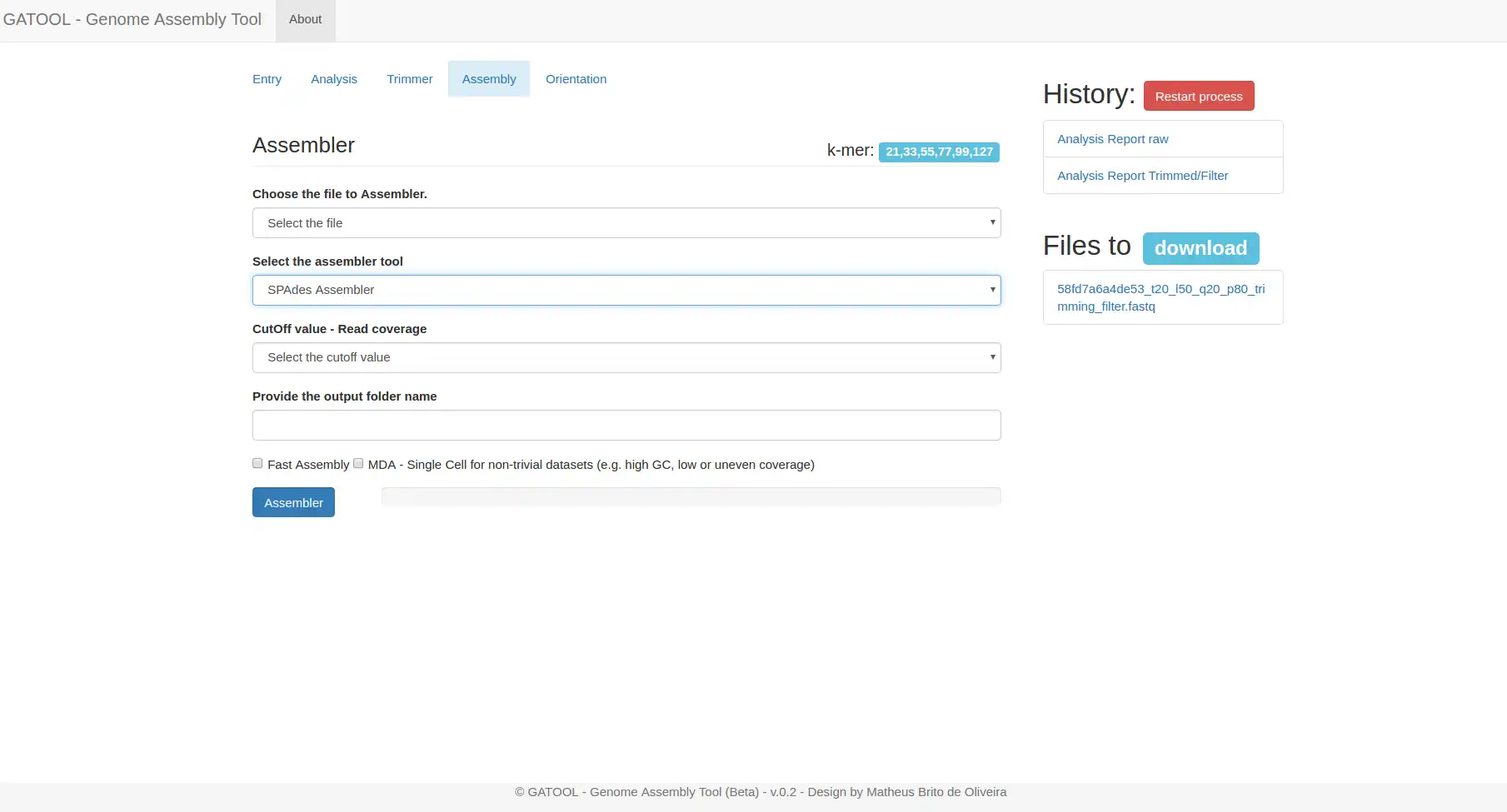
Genome Assembly Tutorial with SPADes | llumina Reads | SPAdes | Bioinformatics for Beginners - YouTube

HGA: de novo genome assembly method for bacterial genomes using high coverage short sequencing reads | BMC Genomics | Full Text